Welcome
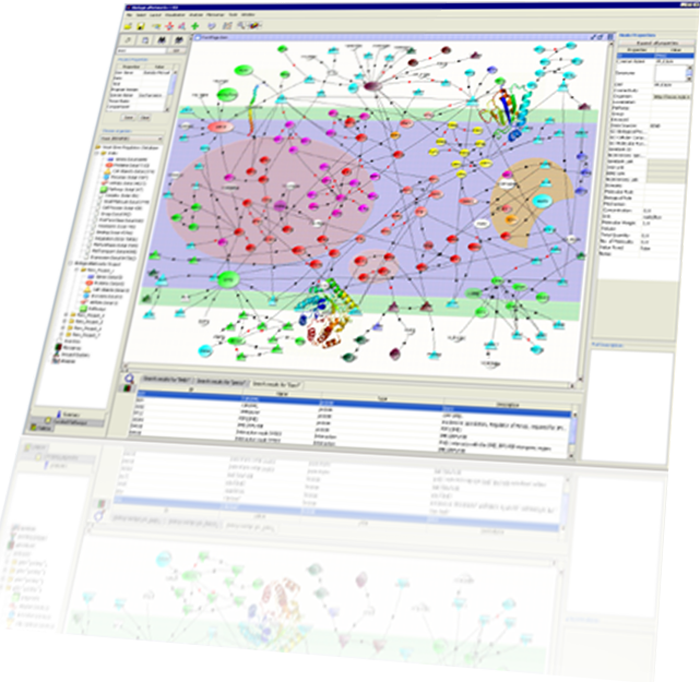
BiologicalNetworks is a bioinformatics and systems biology software platform for visualizing molecular interaction networks, sequence and 3D structure information, integrating these with other graph-structured data such as ontologies (e.g. gene ontology) and taxonomies (e.g. enzyme classification and functional classification of proteins), integrating interactions with experimental data (e.g. gene expression), querying all types of data to extract biologically meaningful relations, dynamical modeling and simulation.
The server provides querying services and an information management framework over PathSys, the system which integrates over 14 curated and publicly contributed data sources for the 8 representative genomes (S. cerevisiae, D.melanogaster, etc. full list see here).



